a) Development of novel BRAF inhibitors (ICB activity)
Biological evaluation is performed of novel putative BRAF pathway inhibitors, designed and synthesized by ICB groups (Goulielmaki et al., 2019). Hit-to-lead optimization. New protocols of drug delivery (Theochari et al., 2017).
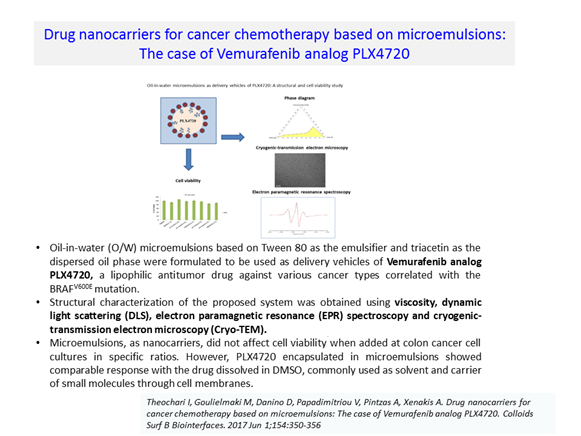
In collaboration with ICB groups for In silico Drug Design, Structural Biology, Synthetic Chemistry, Nanotechnology Goulielmaki M, et al. Transl Oncol. 2019 Jul;12(7):932-950.
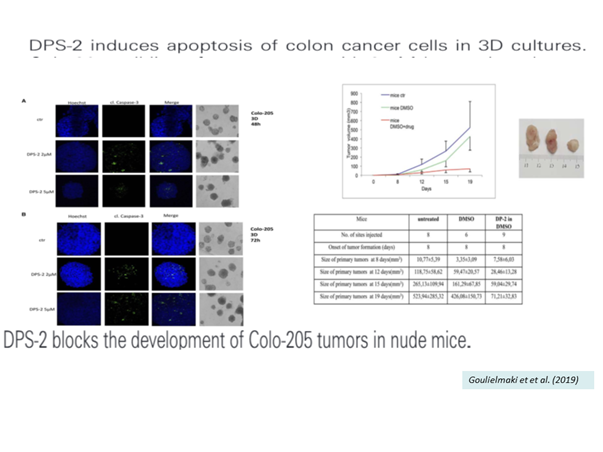
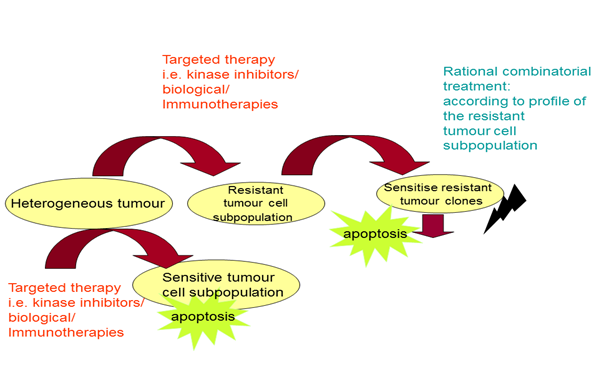
b) Establishment of efficient rational combinations of BRAF/PI3K kinase inhibitors with apoptosis/autophagy modulators to overcome drug resistance in mtBRAF CRC tumours
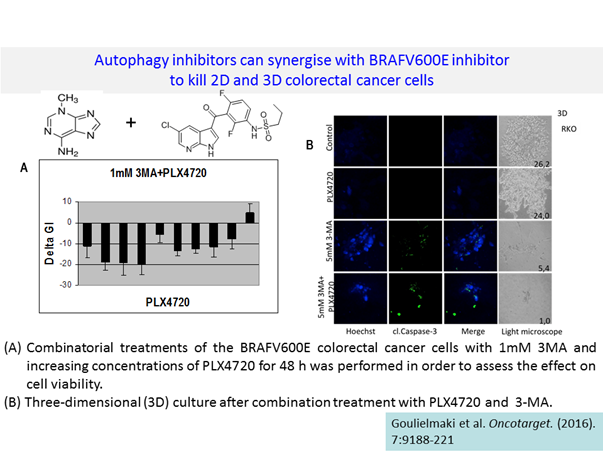
Following the studies on protocols regarding specific BRAF kinase inhibitors with autophagy inhibitors (Goulielmaki et al., 2016); SMAC mimetics with either BRAF/TRAIL/BCl-2 inhibitors (Perimenis et al., 2016):
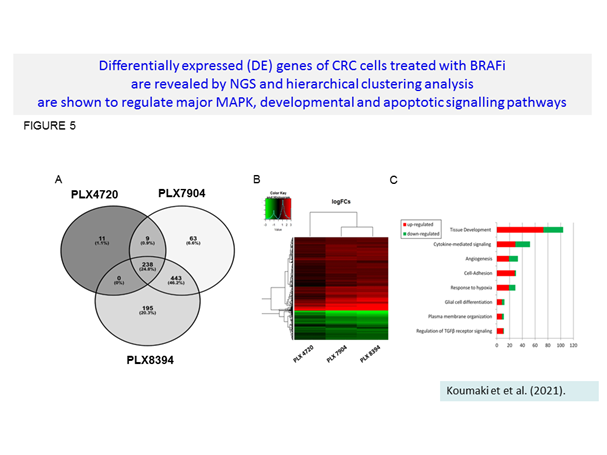
Genomic and bioinformatic analysis revealed the interest for combinatorial treatments of BRAFi with Mcl-1 and Notch modulators show a better effect than the corresponding mono-treatments (Koumaki et al., 2021). This promising combinatorial protocol is further exploited (in preparation, 2022).
Koumaki K, et al. BRAF paradox breakers PLX8394, PLX7904 are more effective against BRAFV600Ε CRC cells compared with the BRAF inhibitor PLX4720 and shown by detailed pathway analysis. Biochim Biophys Acta Mol Basis Dis. 2021 Apr 1;1867(4):166061.
doi: 10.1016/j.bbadis.2020.166061
Supported by the following research grants:
“STHENOS-b”: Targeted therapeutic approaches against degenerative diseases with special focus on cancer and ageing-optimisation of the targeted bioactive molecules” (2017-2021, funding for NHRF/ICB 790kEuros) (co-ordinator); “EATRIS-GR”: Infrastructure for Preclinical and early-Phase Clinical Development of Drugs, Therapeutics and Biomedical Devices (EATRIS-GR) (2019-2022)